Next-Generation Sequencing of Microbial Cell-Free DNA to Rapidly Detect Fluoribacter Bozemanae Pneumonia in an Immunocompromised Host
Raihaan Khattak1, Rameez Rao2, Gurjot Garcha3, Mario Madruga1, Antonio Crespo1, SJ Carlan4*
1Division of Infectious Disease, USA
2Department of Internal Medicine, USA
3Division of Pulmonary and Critical Care Medicine, USA
4Division of Academic Affairs and Research, Orlando Regional Medical Center, Orlando, Florida, USA
*Correspondence author: SJ Carlan, Division of Academic Affairs and Research, Orlando Regional Medical Center, Orlando, Florida, USA;
Email: stevecarlan@gmail.com
Citation: Khattak R, et al. Next-Generation Sequencing of Microbial Cell-Free DNA to Rapidly Detect Fluoribacter Bozemanae Pneumonia in an Immunocompromised Host. Jour Clin Med Res. 2023;4(3):1-4.
Copyright© 2023 by Khattak R, et al. All rights reserved. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
| Received 16 Nov, 2023 | Accepted 01 Dec, 2023 | Published 08 Dec, 2023 |
Abstract
Background: Fluoribacter bozemanae is an uncommon pathogen that accounts for just 3-5% of cases of pneumonia caused by the different Legionella species. The mortality rate has been reported to be up to 40%. This organism is difficult to identify, making diagnosis extremely challenging with current diagnostic modalities.
Case Report: We present a case of a 75-year-old female with a history of multiple myeloma undergoing active treatment with immunosuppressive agents who presented with confusion, hypoxemic respiratory failure and sepsis. Her initial presentation was consistent with Legionella infection, however, all routine testing including a Legionella urine antigen test came back negative. A CT scan of the chest was completed which showed near complete left lower lobe consolidation concerning for pneumonia. An infectious workup was initiated. Atypical and viral pneumonia polymerase chain reaction panel was negative. Microbial cell-free DNA sequencing returned positive for Fluoribacter bozemanae, a subspecies of the Legionellaceae family that is known to cause severe pneumonia in immunocompromised hosts. This information allowed a therapy adjustment resulting in rapid improvement of our patient’s symptoms.
Conclusion: In cases such as this one, the diagnosis of Legionella species is often missed due to the many challenges associated with the different testing modalities. In these circumstances, next-generation genetic sequencing can be extremely useful. Now with the increasing availability of next-generation sequencing as a microbiological diagnostic tool, we can diagnose many more infections that once would have gone undiagnosed and either untreated or delayed in treatment.
Keywords: Legionella; Pneumonia; Microbial Cell-Free DNA Sequencing; Immunocompromised
Article Type
Case Report
Introduction
Over the last decade, there has been a rising interest in Next-Generation Sequencing (NGS) technology as an aid in the diagnosis of microbial infection and thus, in selecting appropriate therapies. The Next Generation Sequencing (NGS) technology can be used to detect the presence of clinically important pathogenic organisms in human specimens [1]. In contrast to human sequencing diagnostics, infectious disease sequencing diagnostics generally require rapid and actionable results, sometimes within hours, as delayed or incorrect initial diagnoses can result in fatalities [1]. We are presenting a clinical scenario in which NGS technology, specifically the Karius test, was used to identify a subspecies of the Legionellaceae family, Fluoribacter bozemanae, causing severe pneumonia in an immunocompromised host [2,3]. The lack of effective commercially available diagnostic modalities for many of the different subspecies and serogroups of the Legionellaceae family poses a unique diagnostic challenge [4]. In our scenario, NGS technology helped us in making a timely diagnosis and allowed us to adjust therapy and achieve a positive clinical outcome.
Case Report
This is a case of a 75-year-old female with a history of multiple myeloma, initially diagnosed the year before, who was actively undergoing treatment with lenalidomide, daratumumab and dexamethasone. She presented to the hospital with chills, weakness, nausea, vomiting and diarrhea. Her symptoms were present for several days and continued to progress rapidly at home.
In the emergency department, she was febrile with a temperature of 103 F and tachycardic with a heart rate of up to 110 and a respiratory rate of 24. Her blood pressure and oxygen saturation were within normal limits. Labs were remarkable for mild hyponatremia with sodium level of 130 mEq/L (normal range: 135 to 145 mEq/L) and elevated erythrocyte sedimentation rate of 105 mm/hr (normal range: 0 to 29 mm/hr) and c-reactive protein of 268 mg/L (normal range: < 10 mg/L). Lactic acid was 1.3 mg/dL (normal range 0.4 to 2.0 mg/dL). Otherwise, her complete blood count, basic metabolic panel, prothrombin time and urine analysis were all normal. A physical exam revealed a dyspneic elderly female with coarse breath sounds noted over the lower left lung field. Initial chest x-ray showed left lower lobe pulmonary infiltrates. She was sepsis alerted and started empirically on intravenous vancomycin and intravenous cefepime. A CT scan of the chest was completed which showed near complete left lower lobe consolidation concerning for pneumonia (Fig. 1). An infectious workup was initiated. Atypical and viral pneumonia polymerase chain reaction panel was negative. Pneumococcal and legionella urinary antigen testing was negative. Bronchoscopy and bronchoalveolar lavage were performed and samples were sent for aerobic, anaerobic, fungal and acid-fast bacilli cultures along with specialized legionella cultures. They all did not show any growth. Legionella polymerase chain reaction testing was not requested at the time.
Due to the lack of improvement despite broad-spectrum antibiotic coverage the Infectious disease team was consulted. They requested the microbial cell-free DNA sequencing test (Karius) from blood. At this point, they were concerned for encapsulated and atypical organisms such as Streptococcus pneumonia, Haemophilus influenza, Moraxella catarrhalis, Staphylococcus aureus, enteric gram-negative rods, Pseudomonas, Legionella sp. and Aspergillus. The cell-free microbial DNA sequencing test resulted within 48 hours and revealed Fluoribacter bozemanae at 45,761 MPMs (molecules per microliter). The patient was subsequently changed to oral Moxifloxacin 400mg daily for 10 days at the recommendation of the infectious disease team and her symptoms improved significantly. She was eventually discharged home in stable condition.
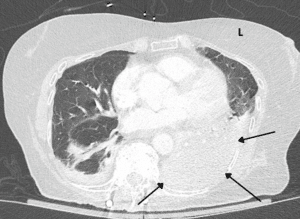
Figure 1: CT chest. The arrows point to the consolidated left lower lobe of the lung.
Discussion
Fluoribacter bozemanae formally known as Legionella bozemanae is formidable species belonging to the family Legionellaceae. It is a small fastidious gram-negative bacillus with special growth requirements which can make diagnosis challenging. Fluoribacter bozemanae is an uncommon pathogen that was first isolated in 1968 [5]. It was named after F. Marilyn Bozeman, the microbiologist who isolated it and first studied the organism. It accounts for just 3-5% of cases of pneumonia caused by Legionella species [6]. Although it is a rare cause of legionellosis which preferentially affects immunocompromised hosts, the mortality rate has been reported to be up to 40% [6,7].
There are currently few rapid diagnostic tests available for the diagnosis of many Legionella spp. and are not widely available. The antigen test in the urine is a common test to detect Legionella infection. The test is specific and accurate even if other methods of testing perform poorly. The urine antigen test performs best when the Pontiac subtype of Lp1 (up to 90%) is the infective agent rather than other monoclonal antibody types of Lp1 (60%) or other Lp serogroups and other species (<5%) [4]. Certain populations of patients such as immunocompromised persons, persons with nosocomial infection or residents of specific geographic locations are more likely to have Legionnaire’s disease caused by another Lp serogroup. Diagnosis caused by these organisms is challenging and other testing modalities are not routinely performed, are user dependent or have low sensitivity or specificity. Some other testing modalities include antibody detection which has low specificity and is an insensitive test in general. It is not routinely recommended especially in serogroups other than Lp 1. Direct fluorescent antibody testing is typically not utilized because it is complex and has a low yield. It also requires a monoclonal antibody to a specific infectious agent [4]. PCR testing plus culture is the gold standard for diagnosis however the sensitivity can vary. Culture sensitivity ranges from anywhere between 20% to 95% and PCR sensitivity ranges from anywhere between 70 to 95% [4]. Unfortunately, there are no commercially approved PCR tests available and tests can only be performed at reference or public health labs. Culture yield heavily depends on the severity of the disease and specific components of the culture media. The standard selective Legionella medium contains cefamandole [7]. However, Fluoribacter bozemanae along with other Legionella serogroups cannot grow on cefamandole-containing media [7]. The diagnosis of Legionnaires disease may be missed unless the culture protocol includes both a nonselective medium and a selective medium that does not contain cefamandole [8]. Consequently, the culture results from many microbiological labs may be inaccurate. Several ancillary tests including PCR, urine antigen test and sputum culture can be used along with urine culture for optimized results.
In cases such as ours, the diagnosis is often missed due to the many challenges associated with the different testing modalities. In these circumstances, next-generation genetic sequencing can be extremely useful. The Karius test successfully diagnosed the patient with Fluoribacter bozemanae pneumonia. The Karius Test is a non-invasive serum test that uses next-generation sequencing of microbial cell-free DNA (mcfDNA) to rapidly detect over 1,400 different bacteria, DNA viruses, fungi and parasites [8]. This test is performed by Karius Inc. There is currently no other validated test that detects pathogen DNA in a small specimen of cell-free plasma like the Karius test [8]. Organisms that present above a predefined statistical significance threshold are reported and quantified in DNA MPM [8]. In our case, the Karius test was able to detect this relatively uncommon yet quite deadly serogroup of Legionella in a timely fashion, allowing us to streamline our antimicrobial therapy and appropriately treat our patient ensuring her recovery.
Conclusion
Fluoribacter bozemanae is a subspecies of the Legionellaceae family which preferentially affects immunocompromised hosts resulting in severe pneumonia. The mortality rate has been reported to be up to 40%. Fluoribacter bozemanae can be difficult to diagnose. Without the use of next-generation sequencing technology, in this case, the organism would not have been detected. The urinary antigen test, although sensitive for Legionella pneumophila serotype I, does not detect many of the other different subspecies and serogroups of the Legionellaceae family. Microbial cultures can be false negative on cefamandole-containing media because Fluoribacter bozemanae cannot grow there. This case highlights the importance of using all diagnostic tools available to clinicians when investigating causes of infection nonresponsive to treatment or unidentifiable with common current lab modalities. It also describes new methods of integrating genetic techniques into human healing.
Conflict of Interest
The authors have no conflict of interest to declare.
References
- Gwinn M, MacCannell D, Armstrong GL. Next-generation sequencing of infectious pathogens. JAMA. 2019;321:893-4.
- Camargo JF, Ahmed AA, Lindner MS. Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000Res. 2019;8:1194.
- Sobel JD, Krieger R, Gilpin R, Griska L, Agarwal P. Legionella bozemanii: still another cause of pneumonia. JAMA. 1983;250:383-5.
- Mercante JW, Winchell JM. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev. 2015;28:95-133.
- Fields BS, Benson RF, Besser RE. Legionella and legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506-26.
- Widmer A, Hohl P, Dirnhofer S, Bassetti S, Marsch S, Frei R. Legionella bozemanii, an elusive agent of fatal cavitary pneumonia. Infection. 2007;35:180-81.
- Lee TC, Vickers RM, Yu VL, Wagener MM. Growth of 28 legionella species on selective culture media: a comparative study. J Clin Microbiol. 1993;31:2764-8.
- Hogan CA, Yang S, Garner OB. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis. 2021;72:239-45.
This work is licensed under a Creative Commons Attribution 2.0 International License.


